Abstract
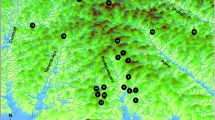
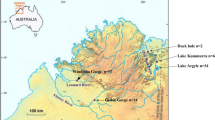
The riverine rabbit, Bunolagus monticularis, is regarded critically endangered. This endemic South African leporid is believed to have extremely low population numbers, and was traditionally thought to be confined to a small region within the Nama Karoo and Succulent Karoo biomes. Recent discoveries of the species at lower altitudes in the Fynbos biome, however, question many of the broadly accepted ideas of the past. The newly defined distribution limits provide an opportunity to assess genetic relatedness across two altitudinally delimited populations providing data that are critical for the development of future conservation efforts, and assessment of the species’ IUCN red list status. We analyzed the degree of geographic genetic structure and mtDNA diversity in Bunolagus using control region sequences from 70 individuals (12 subpopulations) sampled between 1947-2020 across the species’ range. A TCS haplotype network, pairwise AMOVA analysis, and average sequence divergences among subpopulations, all provide genetic support for the recognition of a previously defined Northern and Southern population separated by a semi-permeable geographic barrier comprising unsuitable rocky habitat. Bayesian Skyline analyses link a decline in Bunolagus population numbers to the last glacial maximum but, importantly, the haplotype networks suggest that the Northern and Southern populations responded differently to these paleoclimatic changes. The Northern population, presently confined to the Nama Karoo biome, reflects the effects of fragmentation and survival in refugia in times of increasing aridity, followed by dispersal during more mesic periods. The Southern population, which is mostly confined to Fynbos, exhibits a more stable demographic profile. Surprisingly, given the accepted view of critically low population numbers, Bunolagus exhibits high mtDNA haplotypic diversity underscoring the need for subpopulation connectivity in maintaining genetic diversity through time. We argue that its successful conservation is contingent on reducing human transformation of their habitat and, importantly, protection of sufficient connectivity throughout the species’ range.



Similar content being viewed by others
Availability of Data and Material
All sequence data are available through Genbank – MZ871373 - MZ871442. DNA extracted from the material is housed in the private collection of the Evolutionary Genomics Group at Stellenbosch University, South Africa. Four frozen fibroblasts cells represented by MZ871373 - MZ871376 have been deposited at the National Biobank Facility at the South African National Biodiversity Institute, Pretoria.
References
Abrantes J, Carmo CR, Matthee CA, Yamada F, van der Loo W, Esteves PJ (2011) A shared unusual genetic change at the chemokine receptor type 5 between Oryctolagus, Bunolagus, and Pentalagus. Cons Genet 12:325-330
Adler CJ, Haak W, Donlon D, Cooper A (2011) Survival and recovery of DNA from ancient teeth and bones. J Archaeol Sci 38:956-964
Arbogast BS, Edwards SV, Wakeley J, Beerli P,, Slowinski JB (2002) Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Ann Rev Ecol Syst 33:707–740
Bouckaert R, Vaughan TG, Barido-Sottani J, et al (2019) BEAST 25: An advanced software platform for Bayesian evolutionary analysis. PLoS Comp Biol 15, e1006650
Carr AS, Chase BK, Holmes PJ, Makhubela TV, Rabumbulu M,, Stewart BA (2019) Revisiting the palaeohydrology of the Nama Karoo: late Pleistocene lake high stands and associated archaeology in the South African interior. 20th Congress of the International Union for Quaternary Research (INQUA). doi:1013140/RG223352399363
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG,, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497-3500
Clark PU, Mix AC (2002) Ice Sheets and sea level of the Last Glacial Maximum Quaternary. Sci Rev 21:1–7
Clement M, Snell Q, Walke P, Posada D, Crandall K (2002) TCS: estimating gene genealogies. Proc 16th Int Parallel Distrib Process Symp 2:184
Coates DJ, Byrne M, Moritz C (2018) Genetic diversity and conservation units: Dealing with the species-population continuum in the age of genomics. Front Ecol Evol 23:165
Collins K, du Toit JT (2016) Population status and distribution modelling of the critically endangered riverine rabbit (Bunolagus monticularis). Afr J Ecol 54:195–206
Collins K, Ahlman V, Matthee C, Taylor P, Keith M, Palmer G, van Jaarsveld A (2004) Bunolagus monticularis in Friedmann Y, Daly B, editors Red Data Book of the Mammals of South Africa: A Conservation Assessment IUCN SSC Conservation Breeding Specialist Group and Endangered Wildlife Trust, Johannesburg South Africa
Collins K, Bragg C, Birss C (2019) Bunolagus monticularis The IUCN Red List of Threatened Species 2019: https://www.iucnredlist.org.
de Jong MA, Wahlberg N, van Eijk M, Brakefield PM, Zwaan BJ (2011) Mitochondrial DNA signature for range-wide populations of Bicyclus anynana suggests a rapid expansion from recent refugia. PLoS One 6, e21385
de Menocal PB (2004) African climate change and faunal evolution during the Pliocene-Pleistocene. Earth Planet Sci Let 220:3–24
Dippenaar SM, Ferguson JWH (1994) Towards a captive-breeding programme for the riverine rabbit, Bunolagus monticularis. S Afr J Sci 90:381–385
Duthie AG, Skinner JD, Robinson TJ (1989) The distribution and status of the riverine rabbit Bunolagus monticularis, South Africa. Biol Cons 47:195-202
du Toit N, van Vuuren BJ, Matthee S, Matthee CA (2012) Biome specificity of distinct genetic lineages within the four-striped mouse Rhabdomys pumilio (Rodentia: Muridae) from southern Africa with implications for taxonomy. Mol Phylogenet Evol 65:75-86
Excoffier L, Lischer HEL (2010) Arlequin suite ver 35: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564-567
Edwards S, Claude J, Jansen van Vuuren B, Matthee CA (2011) Evolutionary history of the Karoo bush rat Myotomys unisulcatus (Rodentia: Muridae): disconcordance between morphology and genetics. Biol J Linn Soc 102:510–526
Fitzpatrick BM (2009) Power and sample size for nested analysis of molecular variance. Mol Ecol 18:3961-3966
Hoelzel AR, Hancock JM, Dover GA (1991) Evolution of the cetacean mitochondrial D-Loop region. Mol Biol Evol 8:475–493
Hughes GO, Thuiller W, Midgley GF, Collins K (2008) Environmental change hastens the demise of the critically endangered riverine rabbit (Bunolagus monticularis). Biol Cons 141:23–34
Kalyaanamoorthy S, Minh B, Wong T et al (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589
Kryger U, Robinson TJ, Bloomer P (2004) Population structure and history of southern African scrub hares Lepus saxatilis. J Zool 263:121–133
Lean C, Maclaurin J (2016) The value of phylogenetic diversity In: Pellens R, Grandcolas P (eds) Biodiversity Conservation and Phylogenetic Systematics Topics in Biodiversity and Conservation, Vol. 14. Springer Cham https://doi.org/10.1007/978-3-319-22461-9_2
Leigh JW, Bryant D (2015) PopART: Full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Mable BK (2019) Conservation of adaptive potential and functional diversity: integrating old and new approaches. Conserv Genet 20:89–100
MacPherson AJ, Gillson L,, Hoffman MT (2019) Between- and within-biome resistance and resilience at the fynbos-forest ecotone, South Africa. The Holocene 29:1801-1816
Matthee CA, Robinson TJ (1996) Mitochondrial DNA differentiation among geographical populations of Pronolagus rupestris Smith's red rock rabbit (Mammalia: Lagomorpha). Heredity 76:514–523
Matthee CA, van Vuuren BJ, Bell D, Robinson TJ (2004) A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst Biol 53:433–447
Mead S, Poulter M, Beck J, Uphill J, Jones C, Ang CE, Mein CA, Collinge J (2008) Successful amplification of degraded DNA for use with high-throughput SNP genotyping platforms. Hum Mutat 29:1452-8145
Milot E, Béchet A, Maris V (2020) The dimensions of evolutionary potential in biological conservation. Evol Appl 13:1363–1379
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530-1534
Morgan-Richards M, Bulgarella M, Sivyer L, Dowle EJ, Hale M, McKean NE, Trewick SA (2017) Explaining large mitochondrial sequence differences within a population sample. R Soc Open Sci 4:170730
Mucina L, Rutherford MC (2006) The vegetation of South Africa, Lesotho and Swaziland Strelitzia 19 South African National Biodiversity Institute Pretoria
Nattrass N, Conradie B, Stephens J et al (2020) Culling recolonizing mesopredators increases livestock losses: Evidence from the South African Karoo. Ambio 49:1222–1231
Neparáczki E, Kocsy K, Tóth GE, et al (2017) Revising mtDNA haplotypes of the ancient Hungarian conquerors with next generation sequencing. PLoS ONE 12: e0174886
Osuna F, González D, de los Monteros AE et al (2020) Phylogeography of the volcano rabbit (Romerolagus diazi): the evolutionary history of a mountain specialist molded by the climatic-volcanism interaction in the central mexican highlands. J Mammal Evol 27:745–757
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 17. Syst Biol 67:901-904
Rosel P, Rojas-Bracho L (2006) Mitochondrial DNA variation in the critically endangered vaquita Phocoena sinus Norris and Macfarland 1958. Mar Mamm Sci 15:990-1003
Rouget M, Richardson D M, Cowling R M, Lloyd J W, Lombard A T (2003) Current patterns of habitat transformation and future threats to biodiversity in terrestrial ecosystems of the Cape Floristic Region South Africa. Biol Cons 112:63-85
Rowe RJ, Terry RC (2014) Small mammal responses to environmental change: integrating past and present dynamics. J Mamm 95:1157–1174
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302
Rubinoff D, Holland BS (2005) Between two extremes: Mitochondrial DNA is neither the Panacea nor the Nemesis of phylogenetic and taxonomic inference. Syst Biol 54:952–961
Sands AF, Matthee S, Mfune JK, Matthee CA (2015) The influence of life history and climate driven diversification on the mtDNA phylogeographic structures of two southern African Mastomys species (Rodentia: Muridae: Murinae). Biol J Linn Soc 1141,58–68
Savolainen P, Leitner T, Wilton AN, Matisoo-Smith E,, Lundeberg J (2004) A detailed picture of the origin of the Australian dingo obtained from the study of mitochondrial DNA. Proc Natl Acad Sci U S A 101:12387-12390
Senn H, Banfield L, Wacher T, et al (2014) Splitting or Lumping? A conservation dilemma exemplified by the critically endangered Dama gazelle (Nanger dama) PLoS ONE 9, e98693
Sithaldeen R, Ackermann RR, Bishop JM (2015) Pleistocene aridification cycles shaped the contemporary genetic architecture of southern African baboons. PLoS ONE 10, e0123207
Shields GF, Kocher TD (1991) Phylogenetie relationships of North American ursids based on analysis of mitochondrial DNA. Evolution 45:218–221
Staude IR, Navarro LM, Pereira HM (2020) Range size predicts the risk of local extinction from habitat loss. Global Ecol Biogeogr 29:16–25
Sugden JM (1989) Late quaternary palaeoecology of the central and marginal uplands of the Karoo PhD thesis UCT, South Africa
Urrego DH, Sánchez Goñi1, MF, Daniau AL, Lechevre S, Hanquiez V (2015) Increased aridity in southwestern Africa during the warmest periods of the last interglacial. Clim Past 11:1417–1431
Wang W, Zheng Y, Zhao J et al (2019) Low genetic diversity in a critically endangered primate: shallow evolutionary history or recent population bottleneck? BMC Evol Biol 19:134
Weeks AR, Stoklosa J, Hoffmann AA (2016) Conservation of genetic uniqueness of populations may increase extinction likelihood of endangered species: the case of Australian mammals. Front Zool 13:31
Wogan GOU, Voelker G, Oatley G, Bowie RCK (2020) Biome stability predicts population structure of a southern African aridland bird species. Ecol Evol 10:4066-4081
Zimmerman SJ, Aldridge CL, Oh KP, Cornman RS, Oyler-McCance SJ (2019) Signatures of adaptive divergence among populations of an avian species of conservation concern. Evol Applicat 12:1661–1677
Acknowledgements
Material was provided by landowners and the interested public under auspices and permitting of the Riverine Rabbit Working Group, CapeNature, Northern Cape Environmental Affairs and Nature Conservation, the Amathole Museum, and the Endangered Wildlife Trust Drylands Conservation Programme. Vicky Ahlmann, Bonnie Schumann, Cobus Theron, Guy Palmer, Insauf de Vries, Corne Claasen, Zoe Woodgate, and Jan Humann are especially acknowledged for their sustained support over many years. Daryn Alpers is thanked for performing the DNA extractions on museum specimens. Funding was provided by Stellenbosch University and a National Research Foundation Grant to CAM.
Funding
Funding was provided by Stellenbosch University and a National Research Foundation Grant to CAM.
Author information
Authors and Affiliations
Contributions
CAM: conceptualization, generation of data, data analyses, writing first draft, funding; NDW: generation of data, data analyses, editing of draft, approve final version; TJR: conceptualization, writing and editing of manuscript, approve final version.
Corresponding author
Ethics declarations
Ethics Approval
Material was provided by landowners and the interested public under auspices and permitting of the Riverine Rabbit Working Group, CapeNature, Northern Cape Environmental Affairs and Nature Conservation, the Amathole Museum, and the Endangered Wildlife Trust Drylands Conservation Programme.
Consent to Participate
All authors voluntary participated in conducting this research and had the full approval from Stellenbosch University.
Consent for Publication
All authors read and approved the final version.
Conflicts of Interest/Competing Interests
There are no conflicts of interest or competing interests associated with this work.
Rights and permissions
About this article
Cite this article
Matthee, C.A., de Wet, N. & Robinson, T.J. Conservation Genetics of the Critically Endangered Riverine Rabbit, Bunolagus monticularis: Structured Populations and High mtDNA Genetic Diversity. J Mammal Evol 29, 137–147 (2022). https://doi.org/10.1007/s10914-021-09577-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10914-021-09577-2